Will Grant

Will Grant
Member of Trinity College
PhD student in Dr Ahnert's group
Email: wpg23 @ cam.ac.uk
Personal web site
ORCID: 0000-0001-9309-7937
TCM Group, Cavendish Laboratory
19 JJ Thomson Avenue,
Cambridge, CB3 0HE UK.

Research
The Protein Data Bank now contains over 130,000 solved protein structures, and this number is increasing exponentially. Obtaining the structure of an individual protein yields valuable insight into its function within the cell, and often suggests a mechanism of action which can reveal potential drug targets. However, in order to understand the evolutionary and organisational principles behind protein structures as a whole, the protein structures must be analysed as a set. Abstracting the protein's structure beyond its description in terms of atomic positions may assist in this analysis.
In my current work a protein's structure is converted into a network, in which the atoms or residues of the protein are vertices and a simple distance threshold used to generate edges. This allows the pattern of bonding in the protein to be explored using the pre-existing mathematics of network analysis. The properties of the network can then be mapped onto the properties of the original protein. This method has previously been used to identify key residues in protein dynamics, and in allosteric communication.
One network property that has generated much interest in recent years is that of community structure; the tendency of vertices to form tightly-knit clusters. In my current work, the community structure of protein-based networks is explored. By using pre-existing community detection algorithms, we can find tightly-bound regions of the protein. These regions can then be used to find novel conserved domains, to coarse-grain the protein's structure, and to classify the protein's topology.

My previous work investigated spreading processes (epidemics) on random networks. On the left/below an epidemic on a square lattice within the SIS model is displayed, with Susceptible nodes shown in grey, and Infected nodes in red fading to black.


In Plain English
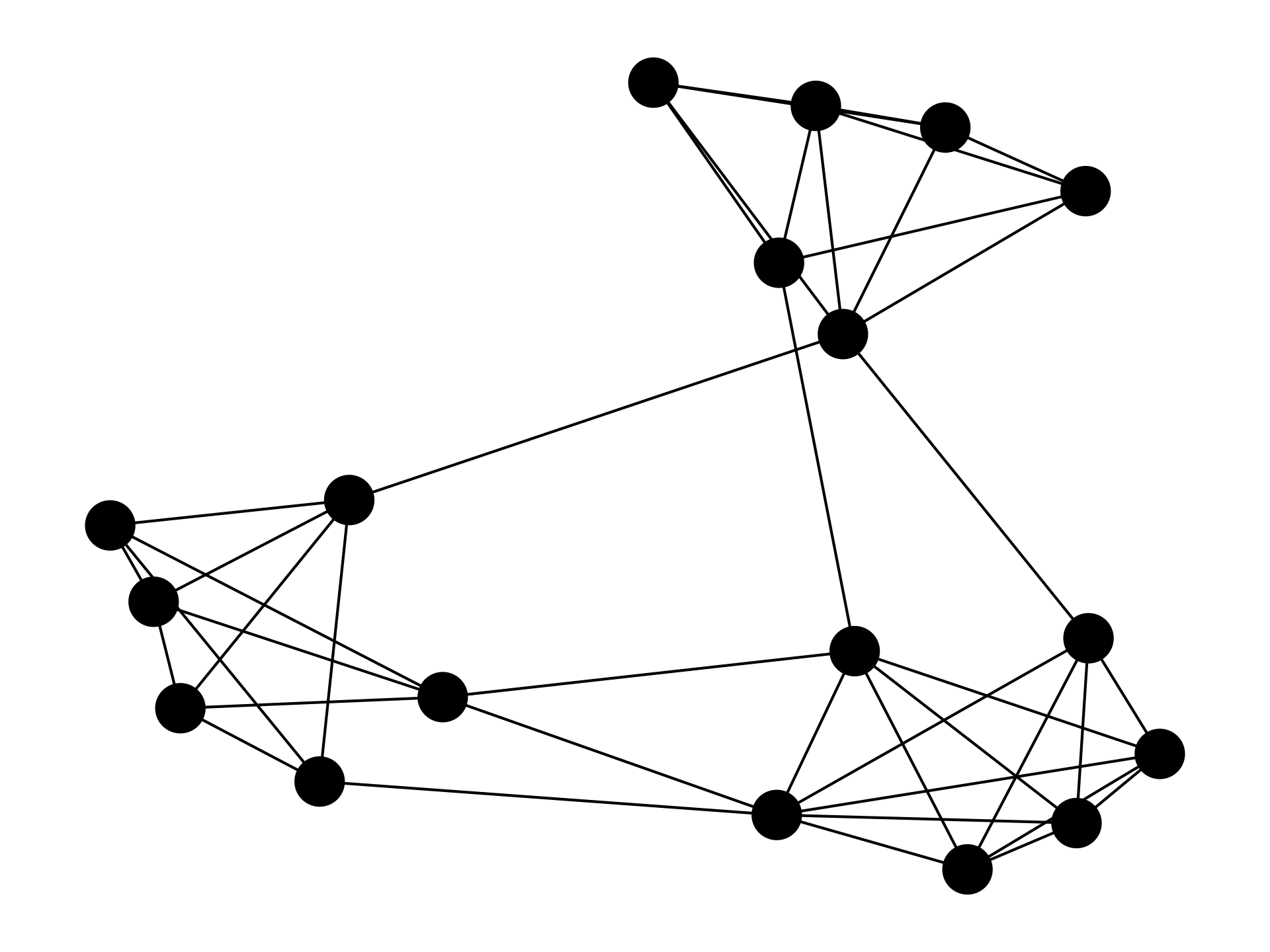
A network is any set of points (known as vertices or nodes) connected by lines (known as edges or links). The network can represent any system with interacting components, from social networks showing people linked by friendship, to computers linked by the Internet.
In my work networks are generated from protein structure. Proteins are folded chains of bonded atoms, and so by creating a network from the atoms of a protein we can acquire a novel view of its bonding pattern.

Featured Publications
- Revealing and exploiting hierarchical material structure through complex atomic networks npj Comput. Mater. 3 UNSP 35 (2017)
- Modular decomposition of protein structure using community detection Journal of Complex Networks (2018)